Restriction-modification systems, abortive-phage phenotypes, toxin-antitoxins and other innate defense systems, in the past, have been shown in familiar chapters in typical microbiology textbook, while now what if I say in prokaryotes world “RISC” can serve a role for new kind of antiviral defense, in addition the “RNAi” can even be engineered and designed to lead to target gene silencing, would you believe me?
You must have ever heard CRISPR/Cas (CRISPR Associated proteins) System if you have ever read this Science paper [1]. Exactly as the title said, CRISPR, Clustered Regularly Inter-spaced Short Palindromic Repeat, serves as the leading role to provide the “memory” as an adaptive immunity, akin to a blacklist of unwanted visitors, like plasmids or viruses genome.
CRISPR/Cas has different types based on Cas family. Three modules of Cas proteins are Cmr, Cst, Csa. It is an old story in bacteria world as it had been firstly identified in E.coli in 1987. Most have been reported to head for invading DNA, while here what I introduce to you now is an unique and intriguing discovery that in achaeon Pyrococcus furiosus which thrives best under extremely high temperatures, CRISPR/Cmr (one subtype of CRISPR/Cas) targets invading RNA, rather than DNA, thus what I called “RNAi” can makes sense.
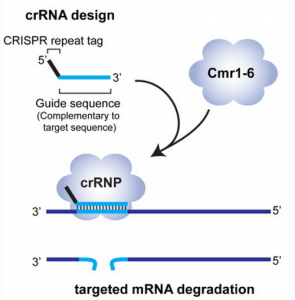
In general, the context of CRISPR RNA (crRNA) is typically a sandwich, repeat-sequence-repeat. The internal sequence is termed guide sequence which is complementary to invading RNA only for CRISPR/Cmr, and it is identical to invading DNA for most other cases of CRISPR/Cas.
–How CRISPR/Cmr works?
 As recalled, the internal sequence of crRNA is complementary to invading RNA as the “seed” region, and more importantly only Cmr complex can contribute to RNA cleavage.
As recalled, the internal sequence of crRNA is complementary to invading RNA as the “seed” region, and more importantly only Cmr complex can contribute to RNA cleavage.
The 8 nt 5′-end tag, among the short repeat sequence in crRNA, will lead crRNA to bind to invading RNA. It is suggested that the 8 nt 5′-end tag plays a discrimination function to classify self-RNA and non-self RNA.
Once crRNA and invading RNA get paired, hydrolysis of the target RNA takes place at a fixed distance, 14nt, from the 3′-end of the small guide RNA. Thus the invading RNA will be degraded and its expression will be turned OFF. In this way Pyrococcus furiosus help themselves against foreign viruses invading with RNA gnome.
Thus I cannot help raising an analogy between CRISPR/Cas system and noted RISC (RNA-induced siliencing complex) in eukaryotes [2]. They all have the progess: processing to be matured, base-pair induced target cleavage.
–Can CRISPR/Cmr engineered?
Yes, we can. The magic is the 8nt 5′-end tag, whose sequence is AUUGAAAG. Scientists had hacked the “immune” system to suppress target gene expression [3], here with the example beta-lactamase (bla) mRNA. The internal sequence, or guide sequence had been designed complementary to 5′-end bla mRNA sequence with the required 5′-end tag. Good result is the gene get silenced which shows promise to another novel silencing systems in bacterium and archaea.
–Questions
CRISPR/Cmr with RNA as its target is just one subtype of CRISPR/Cas system. Other types target DNA.
If we took a deep look at the general features of all the systems, and compare them with eukaryotic RNAi side-by-side,[4] there are still lots of questions remained unsolved, and mechanisms left mysterious.
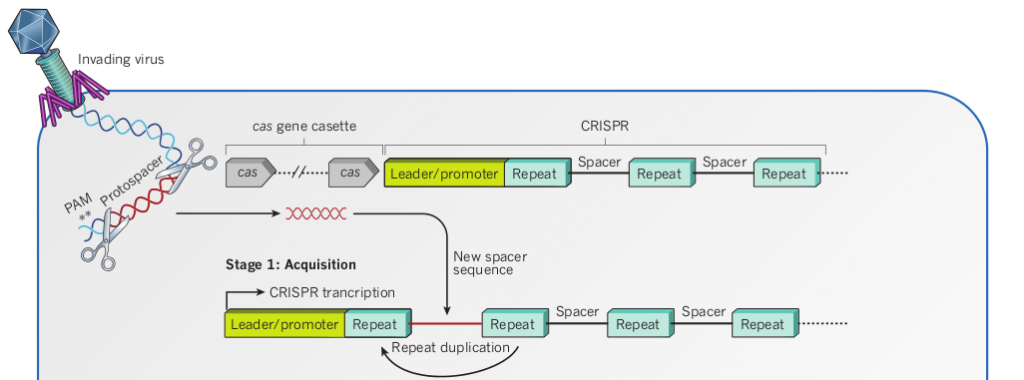
How the invading sequence get integrated into CRISPR loci?
CRISPR/Cas system is adaptive immune system, not innate. Bacteria and archaea are not born with it, and they need immune stimulation at first to gain a short of sequence from invading virus or plasmids. And it is the short foreign sequence that gets integrated into CRISPR loci between two short repeat sequences and enables crRNA to bind with RNA/DNA.
But what is mechanisms of the acquisition? Unknown[4].
How to discriminate self or invading?
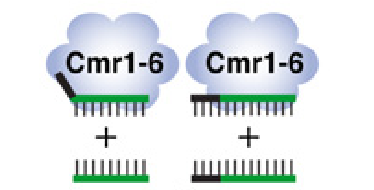
For CRISPR/Cas system that target DNA, the 5′-end tag (in short repeat region) is critical for distinguishing self from non-self. If the 5′-end tag mismatches the invading DNA, the invaders must die. If the tag precisely matches foreign DNA, it is considered as the host CRISPR locus itself and does not “attack”. What about CRISPR/Cmr?
To discern the function of 5′-end tag in CRISPR/Cmr targeting RNA. Three disturbance experiments are conducted. When the 5′-end tag is totally deleted, substituted by other types of sequences (one is precisely complementary to itself, two are with just first one or two bases complementary to itself), these three new tags are no more original 5′-end tag leading to the silencing effect disappear, just as expected. Thus it can be concluded that 5′-end tag is sufficient and critical for RNA silencing.
But an interesting experiment leaves the tag’s function more confusing[3]. If the target transcript sequence is complementary to the tag, even though the target is known to be CRISPR hosts sequence, the RNA cleavage is not prevented. Just like shown in the right half figure, the target sequence cannot escape from being killed even it is complementary to the 5′-end tag. Thus 5′-end seems not to be the key commander in discrimination, or there are other molecules hold the key? At least, another unknown issue.
What can Synbio do?
A DNA silencing systems in bacteria, and novel RNA silencing system in archaea!!! It leaves up to you.
–end &&reference
- Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., Romero, D. A., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science (New York, N.Y.), 315(5819), 1709-12. doi:10.1126/science.1138140
- van der Oost, J., & Brouns, S. J. J. (2009). RNAi: prokaryotes get in on the act. Cell, 139(5), 863-5. doi:10.1016/j.cell.2009.11.018
- Hale, C. R., Majumdar, S., Elmore, J., Pfister, N., Compton, M., Olson, S., Resch, A. M., et al. (2012). Essential Features and Rational Design of CRISPR RNAs that Function with the Cas RAMP Module Complex to Cleave RNAs. Molecular Cell, 1-11. Elsevier Inc. doi:10.1016/j.molcel.2011.10.023
- Wiedenheft, B., Sternberg, S. H., & Doudna, J. a. (2012). RNA-guided genetic silencing systems in bacteria and archaea. Nature, 482(7385), 331-338. doi:10.1038/nature10886
Copyright: The attached figures belong to publications with reference number, respectively.